Rebecca J. Whelan named February 2023 Sutton Family Research Impact Award recipient

The Department of Chemistry congratulates Associate Professor Rebecca J. Whelan on receiving the February 2023 Sutton Family Research Impact Award! Her winning paper was published in ACS Omega and is titled "Simultaneous N-Deglycosylation and Digestion of Complex Samples on S-Traps Enables Efficient Glycosite Hypothesis Generation."
The Sutton Award is a monthly competition among chemistry faculty. Every month, the Chemistry Department Chair and Associate Chairs review the peer-reviewed papers published by chemistry faculty from the three previous months to select a winner. The recipient receives a $500 cash prize and is featured on the departmental website.
For a full list of winners, visit our Sutton Family Research Impact Award webpage.
Simultaneous N-Deglycosylation and Digestion of Complex Samples on S-Traps Enables Efficient Glycosite Hypothesis Generation
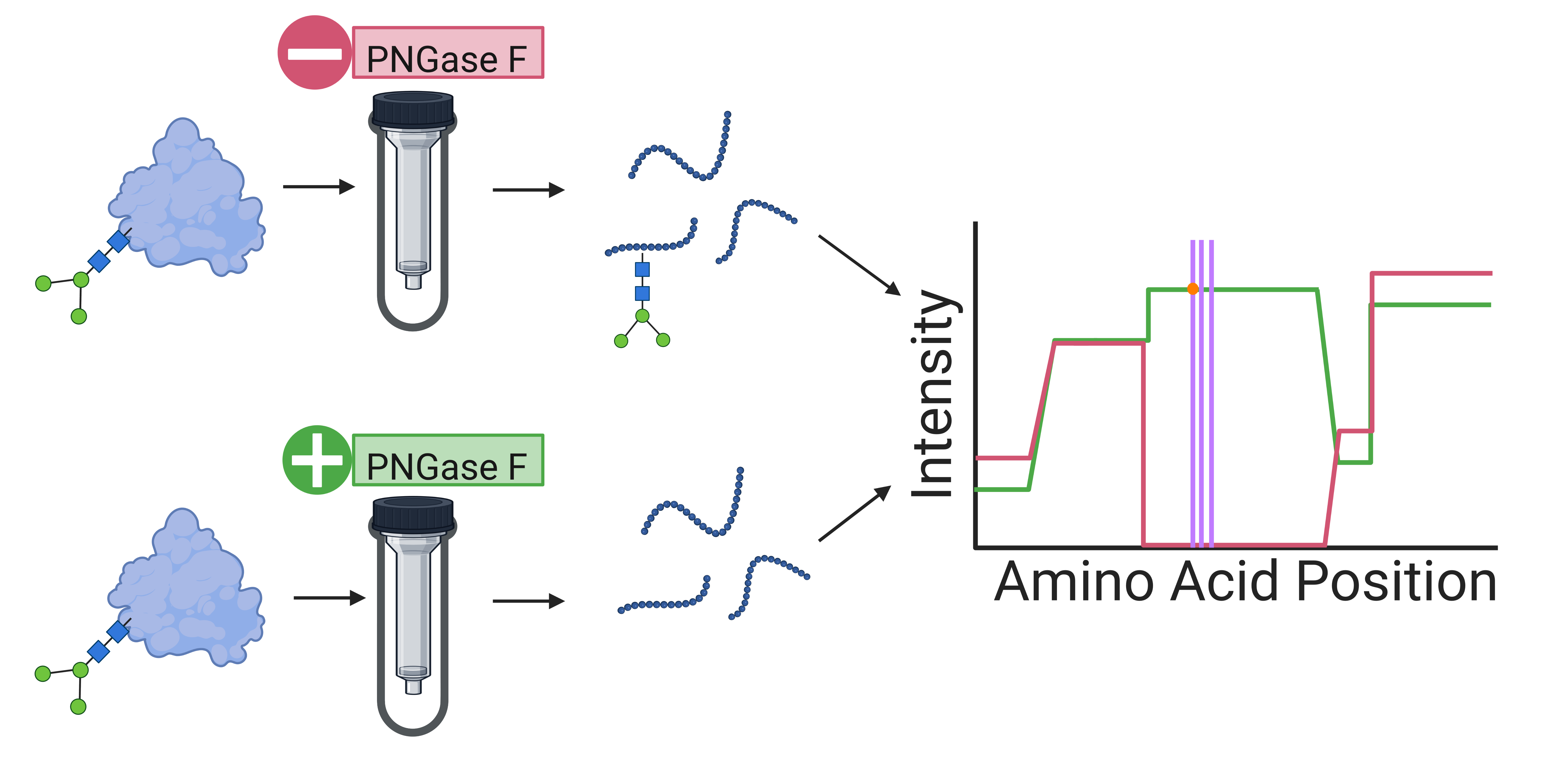
The function of many proteins is modified by the attachment of sugar molecules (“glycans”) at specific locations. One important step in identifying the location and understanding the function of the sugars is removing them so that the protein can be analyzed with and without the sugar modification. Specific enzymes have been shown to be successful in cleaving sugars from proteins prior to protein analysis through a process called deglycosylation, but these enzymatic reactions and subsequent sample clean-up can add up to a day to an already lengthy analysis procedure. An efficient way to integrate deglycosylation with protein analysis would be a valuable contribution. The Whelan Research Group received the February 2023 Sutton Family Research Impact Award for their work on developing a new and highly efficient method of removing sugars from proteins before performing mass-spectrometry-based protein analysis. In their paper, titled “Simultaneous N-Deglycosylation and Digestion of Complex Samples on S-Traps Enables Efficient Glycosite Hypothesis Generation” (DeRosa et al., ACS Omega 2023, 8, 4410-4418), they report the use of suspension trapping (S-Traps), a filtration-based approach, to simultaneously remove N-linked glycans and digest the intact protein into smaller peptides of a size suitable for bottom-up protein analysis.
The investigators applied their method to three different, complex protein mixtures: the complete protein content of bacterial cells, human serum, and excreted proteins produced by ovarian cancer cells. In all cases, the removal of glycans enabled the detection of sections of proteins that were not detectable without glycan removal. Future experiments could use this method to discover potential biomarkers for diseases such as cancer, where aberrant glycosylation is known to play a role. The research team included Professor Whelan, KU Chemistry graduate students Naviya Schuster-Little and Chien-Wei Wang, and two students at the University of Notre Dame, undergraduate Biochemistry major Christine DeRosa and graduate student Simon Weaver, in the Department of Chemistry and Biochemistry and the Integrated Biomedical Sciences Graduate Program.