Professor Steven A. Soper named May 2024 Sutton Family Research Impact Award recipient

The Department of Chemistry congratulates Professor Steven A. Soper on receiving the May 2024 Sutton Family Research Impact Award!
The Sutton Award is a monthly competition among chemistry faculty. Every month, the Chemistry Department Chair and Associate Chairs review the peer-reviewed papers published by chemistry faculty from the three previous months to select a winner. The recipient receives a $500 cash prize and is featured on the departmental website.
For a full list of winners, visit our Sutton Family Research Impact Award webpage.
Detection and identification of single ribonucleotide monophosphates using a dual in-plane nanopore sensor made in a thermoplastic via replication
By Chathurika Rathnayaka, Indu A. Chandrasoma, Junseo Choi, Katie Childers, Maximillian Chibuike, Khurshed Akabirov, Farhad Shiri, Adam R. Hall, Maxwell Lee, Collin McKinney, Matthew Verber, Sunggook Park, and Steven A. Soper
Lab Chip, 2024, 24, 2721-2735
https://doi.org/10.1039/D3LC01062G
DNA sequencing is viewed as a revolutionary technology that is now transitioning into clinical diagnostics to enable precision medicine, which seeks to use genetic information secured from individual patients to improve their care by treating the patient with the correct medications tailored to the molecular characteristics of their disease. However, RNA sequencing is now being realized as the next generation big-data guide to deliver precision care to patients. As an example, patients with breast cancer require that their cancer be molecularly subtyped based on the different types of RNA they express – this information is used to guide the proper treatment regimen. Soper and team are developing a novel RNA sequencing technology that uses a chip-based nanotechnology. In this Lab-on-a-Chip article, the Soper team report a big jump to realize this nanotechnology platform, which consists of the generation of ~8 nm pores fabricated in a plastic via nano-replication. The pores can electrically transduce single molecules in a label-free fashion due to a momentary change in the pore resistance induced by the single molecule (i.e., resistive pulse sensing). Two pores were placed in series with an intervening nanochannel that served as a flight tube to allow the analysis of ribonucleotide molecules, which are the monomer building blocks of RNA (i.e., dual in-plane nanopore sensor). The utility of this dual in-plane nanopore sensor was evaluated to not only detect, but also identify single ribonucleotide monophosphates (rNMPs) by using the travel time (Time-of-Flight, ToF), the resistive pulse event amplitude, and the dwell time. Our data indicated that the identification accuracy of individual rNMPs was high, which was ascribed to an improved chromatographic contribution to the nano-electrophoresis apparent mobility. With the ToF data only, the identification accuracy was 98.3%. However, when incorporating the resistive pulse sensing event amplitude and dwell time in conjunction with the ToF and analyzed via principal component analysis (PCA), the identification accuracy reached 100%. These findings pave the way for the realization of a novel chip-based single-molecule RNA sequencing technology that can easily be transitioned into the clinical laboratory.
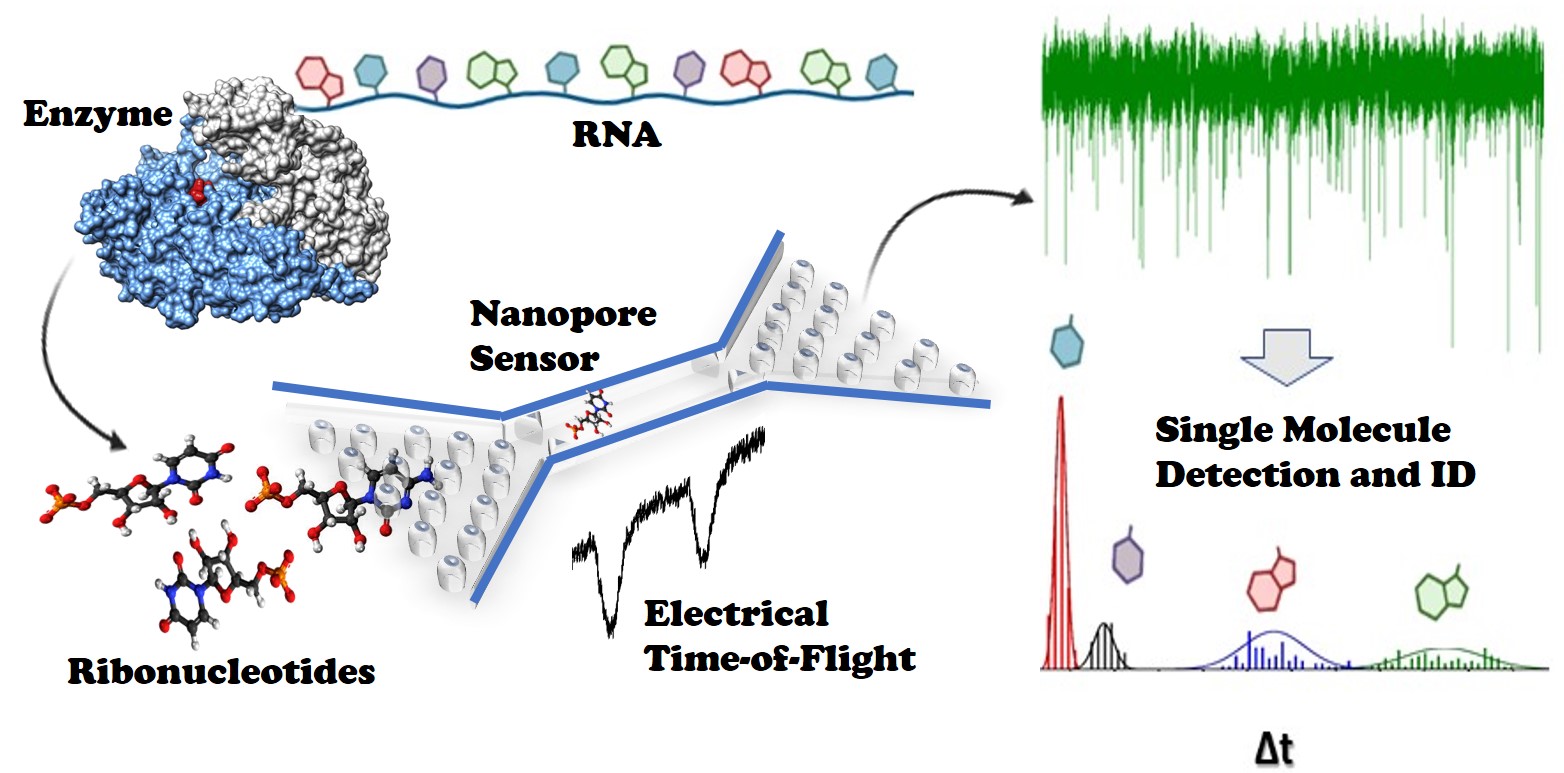
Fig. 1. Abstract figure from paper.

Fig. 2. Dr. Soper and graduate student Indu Chandrasoma in the lab.