Professor Heather Desaire earns January 2022 Sutton Family Research Impact Award

If you’ve been vaccinated against COVID, or if you suffered through a COVID infection, you have had the SARS-CoV-2 spike protein in your body. This protein resides on the surface of the infamous virus and is the molecule that vaccine developers needed to introduce to your immune system, so your body could generate its own defense against SARS-CoV-2, prior to you being exposed.
Heather Desaire’s research group is interested in this protein and the glycans that decorate it. She and Dr. Eden Go, the lead staff scientist in her group, recently published new data analyzing the glycosylation on the spike protein. To accomplish this, they collaborated with two other groups, one at University of Alabama Birmingham, lead by John Kappes, and one at Dana Farber Cancer Center, lead by Joe Sodroski. The Sodroski and Kappes teams worked together to produce well-folded, purified protein to be characterized at KU, where Dr. Go carefully digested the samples then analyzed them using a combination of high-performance liquid chromatography (HPLC) and mass spectrometry.
Winning Paper:
The opportunity cost of automated glycopeptide analysis: case study profiling the SARS-CoV-2 S glycoprotein
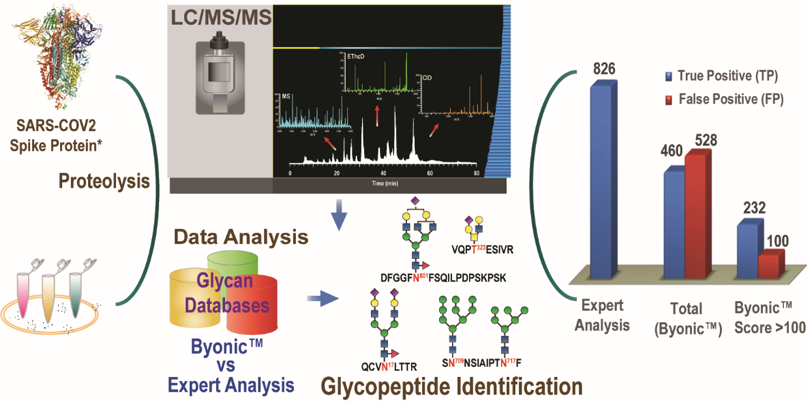
Desaire, Go, and their collaborators are not the first to analyze the glycans on this protein, but they have provided the most detail about which glycans are present, where they reside on the protein, and how diverse the glycosylation profile is at each glycosylation site. This level of detail is necessary for evaluating protein quality when proteins are expressed in vitro for use as pharmaceuticals or vaccines.
The work was initiated during the COVID shut-down, when the Desaire, Kappes, and Sodroski labs had been working on a similar project studying the glycans on a protein under development as an HIV vaccine candidate. The HIV work was put on hold for a short time, when Universities temporarily stopped all research except COVID-related projects. The shut-down drove the research teams to pivot to SARS-CoV-2 work in an effort to use their existing skills to contribute where they could to the COVID crisis.
In addition to generating an in-depth view of the spike protein’s glycosylation profile, the researchers used these data as a test case to evaluate a commercial software product that many researchers in their field had been using to analyze the glycans on this protein and others. This test revealed some rather alarming results: The commercial software produced more false positives (wrong assignments) than true positives (correct assignments), even when it was used under conditions that were advertised to give only 1% false positives. Furthermore, the software was unable to assign hundreds of species that the KU researchers were able to identify and validate. This study serves as a cautionary example that researchers need to be careful about the tools they use to facilitate their work. As it turns out, making software to analyze protein glycosylation is a lot easier than making software that does this task well.