Associate Professor Rebecca Whelan named March 2024 Sutton Family Research Impact Award recipient

The Department of Chemistry congratulates Associate Professor Rebecca Whelan on receiving the March 2024 Sutton Family Research Impact Award!
The Sutton Award is a monthly competition among chemistry faculty. Every month, the Chemistry Department Chair and Associate Chairs review the peer-reviewed papers published by chemistry faculty from the three previous months to select a winner. The recipient receives a $500 cash prize and is featured on the departmental website.
For a full list of winners, visit our Sutton Family Research Impact Award webpage.
A Revised Molecular Model of Ovarian Cancer Biomarker CA125 (MUC16) Enabled by Long-Read Sequencing
By Chien-Wei Wang, Simon D. Weaver, Nicha Boonpattrawong, Naviya Schuster-Little, Manish Patankar, and Rebecca J. Whelan
Published in Cancer Research Communications, 2024, 4, 253–263. https://doi.org/10.1158/2767-9764.CRC-23-0327
Monitoring how patients respond to treatment is a crucial part of the clinical management of ovarian cancer. Currently, patient response is monitored by measuring a protein called CA125 in blood samples. CA125 is an important biomarker of ovarian cancer, and patients and their clinical providers rely on accurate measurements of CA125 concentration. CA125 has been challenging to study, however. At the time CA125 was discovered, the technology that existed for determining the composition of proteins required assembling short sequences, like putting together the pieces of a puzzle. Because of the large size and repetitive nature of CA125, this approach led to gaps and overlaps, incorrectly describing the composition of CA125. Using a technique called Nanopore sequencing, members of the Whelan research group, along with collaborators at the University of Wisconsin and the University of Notre Dame, have been able to determine the sequence of CA125 by reading the entire sequence in one experiment. Compared to the older technology that required piecing many short fragments together, this approach enabled a more complete and accurate molecular model of CA125.
The results of the Nanopore sequencing experiments were corroborated by mass spectrometry experiments in which the CA125 from cultured ovarian cancer cells and tumors from ovarian cancer patients were measured. The mass spectrometry data agree with the model of CA125 enabled by Nanopore sequencing. Finally, the revised model of CA125 was used as the basis of structure prediction in AlphaFold, a deep learning algorithm. The structures predicted by AlphaFold reveal locations on the protein likely to be detected in the clinical test. This aspect of the study is significant because the structure of CA125 has historically been difficult to characterize by conventional methods such as NMR and x-ray diffraction.
The results reported in this article, which was recognized with the March 2024 Sutton Award, lay a foundation for further characterization of this important ovarian cancer biomarker. Having an accurate, revised molecular model of CA125 will enable the Whelan lab and other researchers to improve the existing tests used to monitor ovarian cancer patients’ response to treatment and develop new strategies to better support ovarian cancer clinical care.
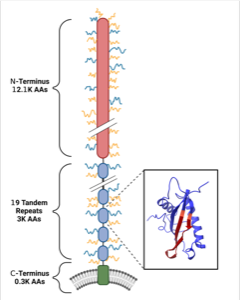
Figure 1. Revised molecular model of CA125 proposed in this study. The model includes: a highly glycosylated N-terminal domain, a 19-unit tandem repeat domain that contains the regions detected in the clinical test, and a short C-terminal domain that includes a cell-membrane spanning region. The inset shows a representative 3D structure of one tandem repeat domain, predicted by AlphaFold. Figure created with Biorender.

Figure 2. Dr. Naviya Schuster-Little (left), Prof. Rebecca Whelan (middle), and Chien-Wei Wang (right) were the KU-based authors on the March 2024 Sutton Award winning article. Naviya will begin a postdoctoral appointment at the University of Wisconsin-Madison in June 2024, and Chien-Wei will be spending the summer as a mass spectrometry method development intern at Genentech.